As title suggests, lets get is going…
Create the Conda Environment with Python 3.5
$ conda create -n python35 python=35
$ conda activate python35
Verify the Conda Environment with python 3.5
$ python
Python 2.7.14 |Anaconda custom (64-bit)| (default, Dec 7 2017, 11:07:58)
Now we will install tensorflow latest which will install lots of required dependency I really needed:
$ conda install -c conda-forge tensorflow
Python run time environment and Folder
Now we will look to confirm the python path
$ which python
/Users/avkashchauhan/anaconda3/bin/python
Now we need to find out where the Python.h header file is which will be used as the values for PYTHON3_INCLUDE_DIR later:
$ ll /Users/avkashchauhan/anaconda3/envs/python35/include/python3.5m/Python.h
Now we need to find out where the libpython3.5m.dylib library file is which will be used as the values forPYTHON3_LIBRARY later:
$ ll /Users/avkashchauhan/anaconda3/envs/python35/lib/libpython3.5m.dylib
Lets clone the OpenCV master repo and opencv_contrib at the same base folder and as below:
$ git clone https://github.com/opencv/opencv
$ git clone https://github.com/opencv/opencv_contrib
Lets create the build environment:
$ cd opencv
$ mkdir build
$ cd build
Now Lets configure the build environment first:
$ cmake -D CMAKE_BUILD_TYPE=RELEASE \
-D CMAKE_INSTALL_PREFIX=/usr/local \
-D OPENCV_EXTRA_MODULES_PATH=../../opencv_contrib/modules \
-D PYTHON3_LIBRARY=/Users/avkashchauhan/anaconda3/envs/python35/lib/libpython3.5m.dylib \
-D PYTHON3_INCLUDE_DIR=/Users/avkashchauhan/anaconda3/envs/python35/include/python3.5m/ \
-D PYTHON3_EXECUTABLE=/Users/avkashchauhan/anaconda3/envs/python35/bin/python \
-D BUILD_opencv_python2=OFF \
-D BUILD_opencv_python3=ON \
-D INSTALL_PYTHON_EXAMPLES=ON \
-D INSTALL_C_EXAMPLES=OFF \
-D BUILD_EXAMPLES=ON ..
The configuration shows following key settings:
......
......
-- Found PythonInterp: /Users/avkashchauhan/anaconda3/bin/python2.7 (found suitable version "2.7.14", minimum required is "2.7")
-- Could NOT find PythonLibs: Found unsuitable version "2.7.10", but required is exact version "2.7.14" (found /usr/lib/libpython2.7.dylib)
-- Found PythonInterp: /Users/avkashchauhan/anaconda3/envs/python35/bin/python (found suitable version "3.5.4", minimum required is "3.4")
-- Found PythonLibs: YYY (Required is exact version "3.5.4")
....
-- Python 3:
-- Interpreter: /Users/avkashchauhan/anaconda3/envs/python35/bin/python (ver 3.5.4)
-- Libraries: YYY
-- numpy: /Users/avkashchauhan/anaconda3/envs/python35/lib/python3.5/site-packages/numpy/core/include (ver 1.12.1)
-- packages path: lib/python3.5/site-packages
--
-- Python (for build): /Users/avkashchauhan/anaconda3/bin/python2.7
-- Pylint: /Users/avkashchauhan/anaconda3/bin/pylint (ver: 1.8.2, checks: 116)
--
General configuration for OpenCV 3.4.1-dev =====================================
-- Version control: 3.4.1-26-g667f5b655
Building the OpenCV code:
Now lets build the code:
$ make -j4
The successful build output end with the following console log:
Scanning dependencies of target example_face_facemark_demo_aam
[ 99%] Building CXX object modules/face/CMakeFiles/example_face_facemark_demo_aam.dir/samples/facemark_demo_aam.cpp.o
[ 99%] Linking CXX executable ../../bin/example_face_facemark_lbf_fitting
[ 99%] Built target example_face_facemark_lbf_fitting
[ 99%] Building CXX object modules/face/CMakeFiles/opencv_test_face.dir/test/test_facemark_lbf.cpp.o
[ 99%] Linking CXX executable ../../bin/example_face_facerec_save_load
[ 99%] Built target example_face_facerec_save_load
[ 99%] Building CXX object modules/face/CMakeFiles/opencv_test_face.dir/test/test_loadsave.cpp.o
[100%] Building CXX object modules/face/CMakeFiles/opencv_test_face.dir/test/test_main.cpp.o
[100%] Linking CXX executable ../../bin/example_face_facemark_demo_aam
[100%] Built target example_face_facemark_demo_aam
[100%] Linking CXX executable ../../bin/opencv_test_face
[100%] Built target opencv_test_face
Lets install is locally:
To install the final library try the following:
$ sudo make install
Once install is completed you will confirm the build output as below:
$ ll /usr/local/lib/python3.5/site-packages/cv2.cpython-35m-darwin.so
Copying final openCV library to Python 3.5 site package:
As we know that Python 3.5 Conda environment folder site-packages is here:
/Users/avkashchauhan/anaconda3/envs/python35/lib/python3.5/site-packages
So we will copy to final cv2.cpython-35m-darwin.so to Python 3.5 Conda environment folder site-packages as cv2.so as below:
$ cp /usr/local/lib/python3.5/site-packages/cv2.cpython-35m-darwin.so
/Users/avkashchauhan/anaconda3/envs/python35/lib/python3.5/site-packages/cv2.so
Confirm it:
$ ll /Users/avkashchauhan/anaconda3/envs/python35/lib/python3.5/site-packages/cv2.so
Verification OpenCV with Python 3.5:
Now Verify the OpenCV with Python 3.5 on Conda Environment:
$ python
Python 3.5.4 |Anaconda, Inc.| (default, Feb 19 2018, 11:51:41)
[GCC 4.2.1 Compatible Clang 4.0.1 (tags/RELEASE_401/final)] on darwin
Type "help", "copyright", "credits" or "license" for more information.
>>> import cv2
>>> cv2.__version__
'3.4.1-dev'
>>>
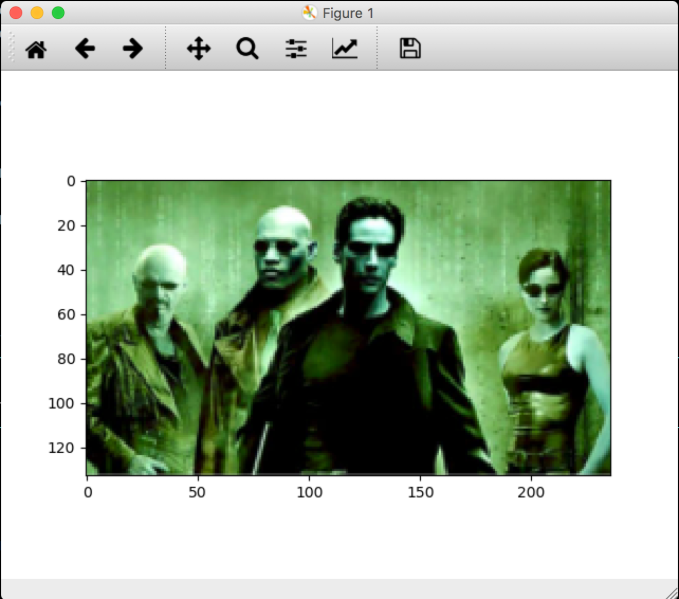
Now lets run OpenCV with as example:
import numpy as np
import cv2
# Load an color image in grayscale
img = cv2.imread('/work/src/github/aiprojects/avkash_cv/test_image.png', 0)
while(True):
cv2.startWindowThread()
cv2.namedWindow("preview")
# Display the resulting frame
cv2.imshow("preview", img)
if cv2.waitKey(1) & 0xFF == ord('q'):
break
# When everything done, release the capture
cv2.destroyAllWindows()
Thats it, enjoy!!
@avkashchauhan