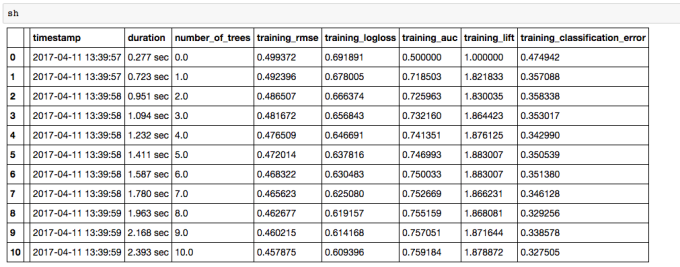
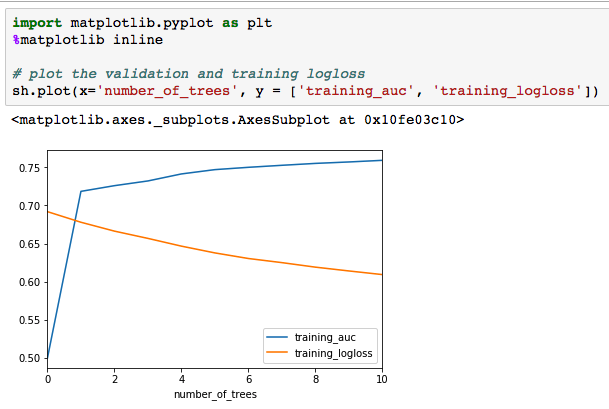
Here is Scala code for binomial classification with GLM:
https://aichamp.wordpress.com/2017/04/23/binomial-classification-example-in-scala-and-gbm-with-h2o/
To add cross validation you can do the following:
def buildGLMModel(train: Frame, valid: Frame, response: String)
(implicit h2oContext: H2OContext): GLMModel = {
import _root_.hex.glm.GLMModel.GLMParameters.Family
import _root_.hex.glm.GLM
import _root_.hex.glm.GLMModel.GLMParameters
val glmParams = new GLMParameters(Family.binomial)
glmParams._train = train
glmParams._valid = valid
glmParams._nfolds = 3 ###### Here is cross-validation ###
glmParams._response_column = response
glmParams._alpha = Array[Double](0.5)
val glm = new GLM(glmParams, Key.make("glmModel.hex"))
glm.trainModel().get()
}
To look cross-validated model try this:
scala> glmModel._output._cross_validation_models
res12: Array[water.Key[_ <: water.Keyed[_ <: AnyRef]]] =
Array(glmModel.hex_cv_1, glmModel.hex_cv_2, glmModel.hex_cv_3)
Now to get each model do the following:
scala> val m1 = DKV.getGet("glmModel.hex_cv_1").asInstanceOf[GLMModel]
And you will see the following:
scala> val m1 = DKV.getGet("glmModel.hex_cv_1").asInstanceOf[GLMModel]
m1: hex.glm.GLMModel =
Model Metrics Type: BinomialGLM
Description: N/A
model id: glmModel.hex_cv_1
frame id: glmModel.hex_cv_1_train
MSE: 0.14714406
RMSE: 0.38359362
AUC: 0.7167627
logloss: 0.4703465
mean_per_class_error: 0.31526923
default threshold: 0.27434438467025757
CM: Confusion Matrix (vertical: actual; across: predicted):
0 1 Error Rate
0 10704 1651 0.1336 1,651 / 12,355
1 1768 1790 0.4969 1,768 / 3,558
Totals 12472 3441 0.2149 3,419 / 15,913
Gains/Lift Table (Avg response rate: 22.36 %):
Group Cumulative Data Fraction Lower Threshold Lift Cumulative Lift Response Rate Cumulative Response Rate Capture Rate Cumulative Capture Rate Gain Cumulative Gain
1 0.01005467 0....
scala> val m2 = DKV.getGet("glmModel.hex_cv_2").asInstanceOf[GLMModel]
m2: hex.glm.GLMModel =
Model Metrics Type: BinomialGLM
Description: N/A
model id: glmModel.hex_cv_2
frame id: glmModel.hex_cv_2_train
MSE: 0.14598908
RMSE: 0.38208517
AUC: 0.7231473
logloss: 0.46717605
mean_per_class_error: 0.31456697
default threshold: 0.29637953639030457
CM: Confusion Matrix (vertical: actual; across: predicted):
0 1 Error Rate
0 11038 1395 0.1122 1,395 / 12,433
1 1847 1726 0.5169 1,847 / 3,573
Totals 12885 3121 0.2025 3,242 / 16,006
Gains/Lift Table (Avg response rate: 22.32 %):
Group Cumulative Data Fraction Lower Threshold Lift Cumulative Lift Response Rate Cumulative Response Rate Capture Rate Cumulative Capture Rate Gain Cumulative Gain
1 0.01005873 0...
scala> val m3 = DKV.getGet("glmModel.hex_cv_3").asInstanceOf[GLMModel]
m3: hex.glm.GLMModel =
Model Metrics Type: BinomialGLM
Description: N/A
model id: glmModel.hex_cv_3
frame id: glmModel.hex_cv_3_train
MSE: 0.14626761
RMSE: 0.38244948
AUC: 0.7239823
logloss: 0.46873763
mean_per_class_error: 0.31437498
default threshold: 0.28522220253944397
CM: Confusion Matrix (vertical: actual; across: predicted):
0 1 Error Rate
0 10982 1490 0.1195 1,490 / 12,472
1 1838 1771 0.5093 1,838 / 3,609
Totals 12820 3261 0.2070 3,328 / 16,081
Gains/Lift Table (Avg response rate: 22.44 %):
Group Cumulative Data Fraction Lower Threshold Lift Cumulative Lift Response Rate Cumulative Response Rate Capture Rate Cumulative Capture Rate Gain Cumulative Gain
1 0.01001182 0...
scala>
Thats it, enjoy!!